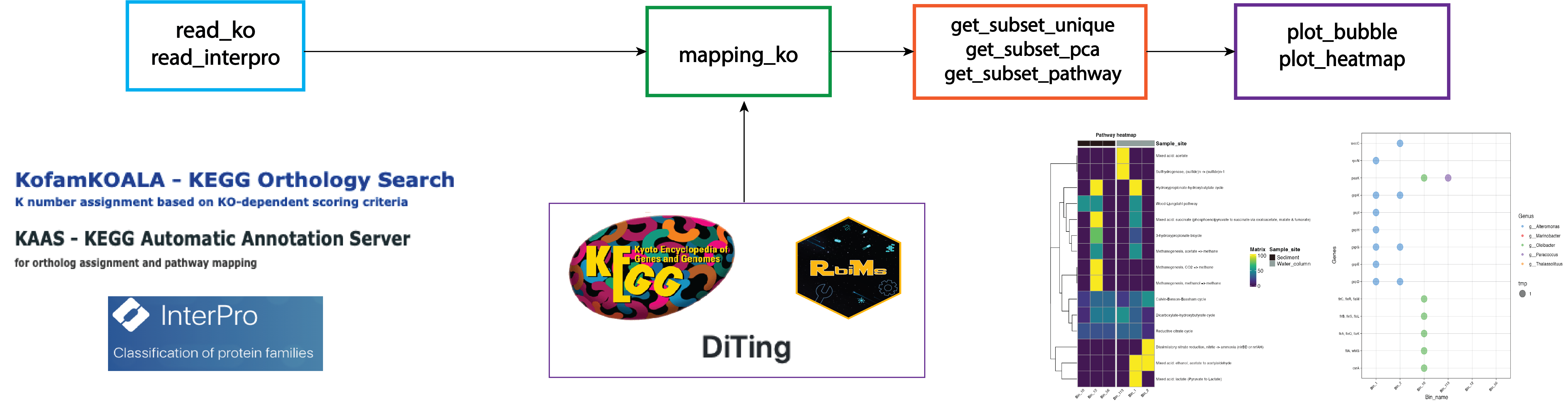
R tools for reconstructing bin metabolisms.
Quick install
We are actively developing rbims, so for now it is not available at CRAN or Bioconductor, however you can access the development version from GitHub.
In R terminal:
install.packages("devtools")If you are in a MAC system, you will need to download XQuartz, you can read more about it here. As well if you are in ubuntu you need to install libcairo2-dev, you can read more about it here.
library(devtools)
install_github("mirnavazquez/RbiMs")
library(rbims)Visit the website for more detail.
New function to write a metabolism workbook
If you want to create a metabolism table in excel that includes KEGG and interproscan outputs you can use this new function.
library(rbims)
write_metabolism("Interpro_test.tsv",
"path/to/KEGG/output/")Contributions
- Mirna Vázquez Rosas Landa - wrote the code.
- Kathryn Appler, Valerie De Anda and Pedro Leão - Curated the rbims database of metabolism.
- Sahil Shah - Helped with function documentation.
- Bryan Vernon - Editor and shiny app developer.
References
- Kanehisa, M. and Goto, S.; KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27-30 (2000).
- Kanehisa, M; Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947-1951 (2019).
- Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M., and Tanabe, M.; KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545-D551 (2021).
- DiTing cycles definition.